242-复现果蝇ChIP-Seq文章的IGV结果
刘小泽写于2021.4.21 这里有两个IGV小技巧,可以学习一下
文章简介
文章是2018年elife的:Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila(DOI: https://doi.org/10.7554/eLife.35368.001)
目的:To explore the mechanism by which age-modulated increase of H3K27me3 impacts adult lifespan
发现: epigenetic drift of H3K27me3 is one of the molecular mechanisms that contribute to aging and that stimulation of glycolysis promotes metabolic health and longevity
要复现的图是
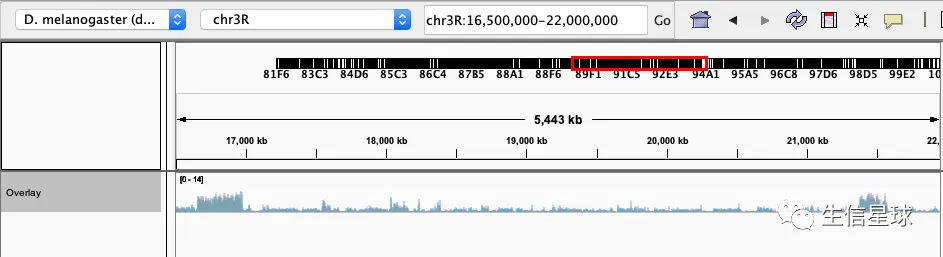
figure 1 F,其中
- 3d (grey), 15d (cyan), and 30d (pink) (top panel)
- deducting 3d signals from those at 15d (middle panel)
- deducting 15d signals from those at 30d of age (bottom panel).
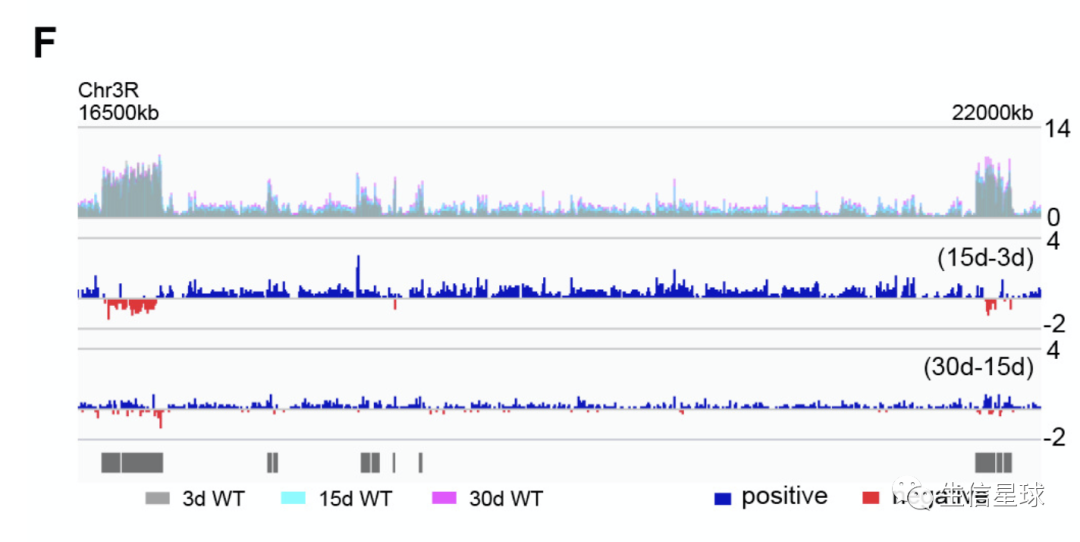
因此,需要拿到bw文件后,做一个重叠效果,然后做两个bw相减的效果
首先拿到bw文件
数据在:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96654
我们只需要下载三个文件:
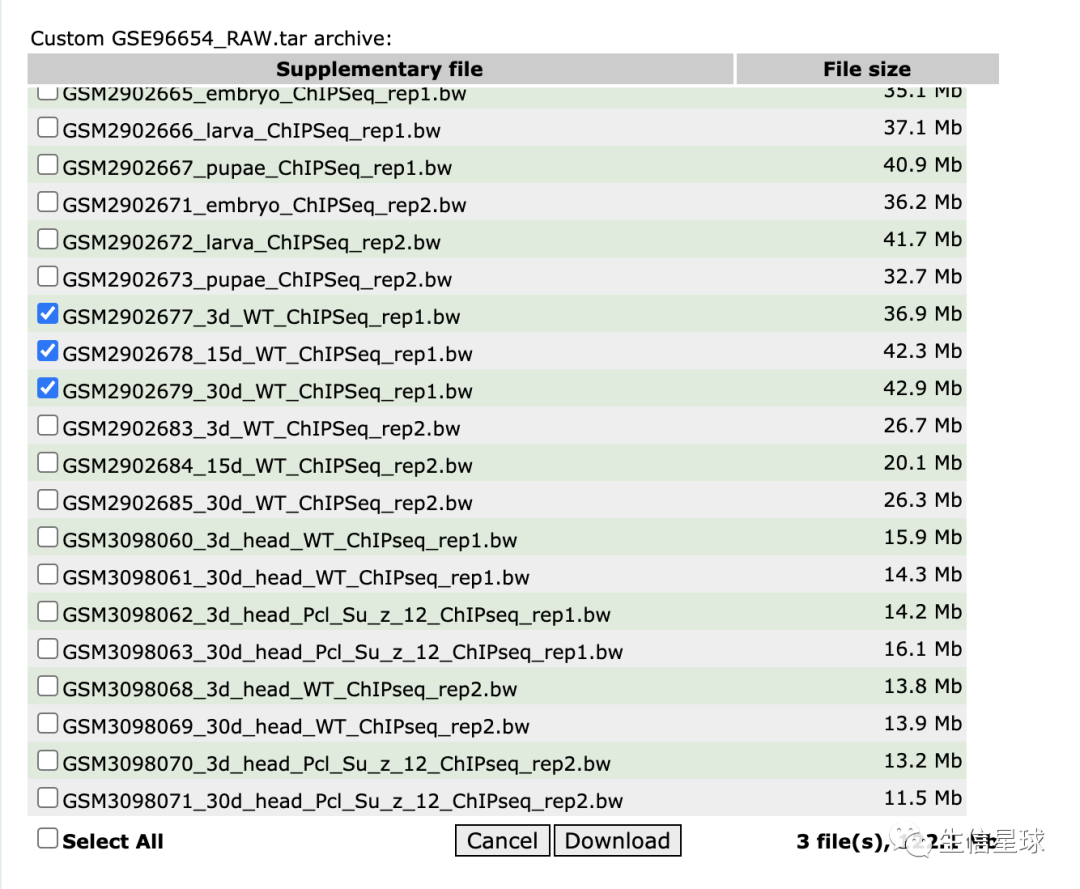
然后打开IGV,导入数据
这里参考基因组是果蝇的,然后按文章描述定位到:chr3R:16,500,000-22,000,000
并且调整好颜色:Change Track Color
然后设置统一的数据范围:Set Data Range 为 0-14
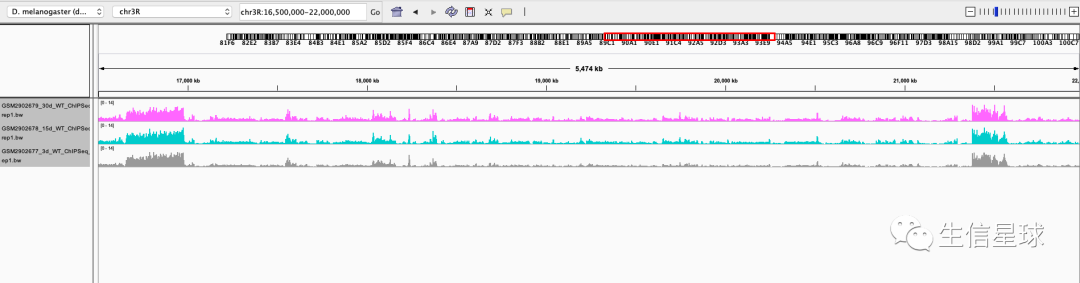
然后看到有一个选项Overlay Tracks:
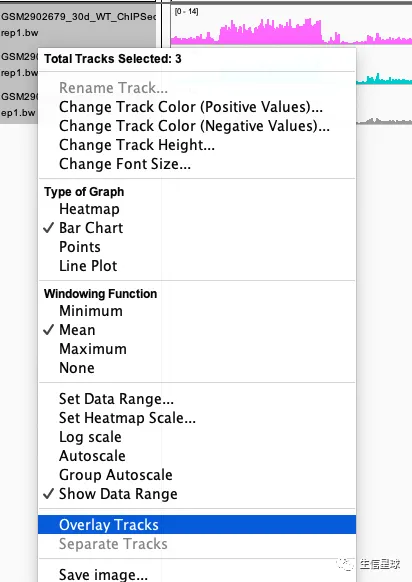
并且可以通过Adjust Transparency调整透明度,得到的结果就是:
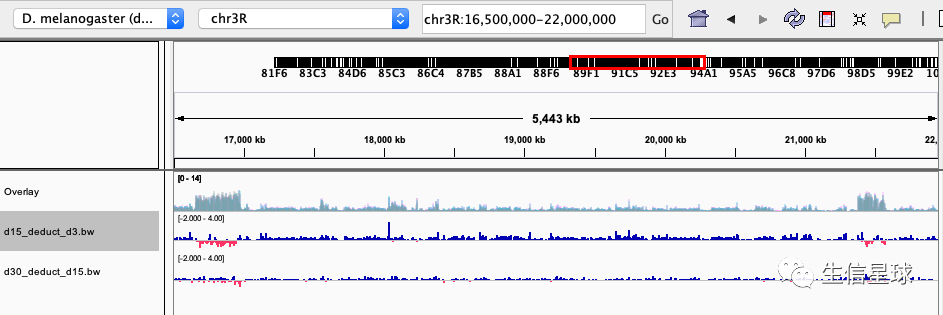
然后进行bw deduct操作
这个可以借助deeptools,帮助文档在:https://deeptools.readthedocs.io/en/develop/content/tools/bigwigCompare.html
其中有个参数--operation ,它可以包括:log2, ratio, subtract, add, mean, reciprocal_ratio, first, second
# d15-d3
# Bigwig file 1. Usually the file for the treatment.
# Bigwig file 2. Usually the file for the control.
bigwigCompare --bigwig1 GSM2902678_15d_WT_ChIPSeq_rep1.bw --bigwig2 GSM2902677_3d_WT_ChIPSeq_rep1.bw --operation subtract --outFileName d15_deduct_d3.bw -p 4
# d30-d15
bigwigCompare --bigwig1 GSM2902679_30d_WT_ChIPSeq_rep1.bw --bigwig2 GSM2902678_15d_WT_ChIPSeq_rep1.bw --operation subtract --outFileName d30_deduct_d15.bw -p 4
拿到这两个bw后,导入IGV,并且设置范围是-2 <=> 4
或者还可以直接利用IGV tools:Tools => Combind tracks